Introduction
Sepsis is a life-threatening infection with a mortality rate above 25%. Rapid identification of the microorganisms that cause bloodstream infections is important for choosing targeted and timely antimicrobial treatment (1). The implementation of matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) technology in laboratories has accelerated microbial diagnosis time (2).
Nevertheless, according to the conventional algorithm, 16-24 hours are needed for pathogen identification and at least an additional 24 hr for antimicrobial susceptibility testing (AST) results. There are several rapid culture-independent techniques to detect bloodstream pathogens, but these are expensive for most diagnostic laboratories in developing countries (3). Some in-house extraction methods and commercial kits have been developed to obtain bacterial pellets directly from positive blood cultures. Identification can be carried out using MALDI-TOF MS, automated identification (ID)/AST systems, or biochemical tests via bacterial pellets. However, they require manual processing time for lysis and centrifugation steps, additional personnel, and cost (4). Therefore, despite technological developments, implementing these methods in routine laboratory workflows is challenging for many laboratories.
In previous years, a novel method for the rapid identification of microorganisms from positive blood culture (BC) bottles based on MALDI-TOF MS analysis after decreased incubation time on a solid medium was introduced (5). In this study, we evaluated this mass spectrometry-based approach for the identification of bacteria from positive BCs and analyzed the method’s performance at short-term incubation times.
Materials and Methods
Positive BC bottles (162 BACTEC™ Plus/F Aerobic and 81 BACTEC™ Lytic Anaerobic bottles; BD Diagnostics, USA) were examined over a period of five months (from January 01, 2021, to May 31, 2021) in this study. For positive BC bottles from the same patient, only the initial bottle flagged as positive was included in the study. The BC bottles were incubated in an automated BC system (BACTECTM FX; BD Diagnostics, USA). Following the positive signal, the Gram staining was performed, and all samples were inoculated onto the Columbia blood agar, chocolate agar, and MacConkey agar. Anaerobic bottles were also inoculated onto the Schaedler agar. The plates were incubated at 36±1 °C in the air with 5% CO2. Schaedler agars were incubated in the anaerobic jar (Oxoid Ltd., United Kingdom) containing GasPak (Thermo Fisher Scientific, USA). MALDI-TOF MS was performed at time points of 2 hr, 4 hr, and 6 hr from immature biomass grown on solid media until successful identification was achieved.
The analysis was done using the Microflex LT system (Bruker Daltonics, Germany) and MALDI BIOTYPER 3.3 (Bruker Daltonics) software. If the score was ≥2.0, the microorganism is identified at the species level and ≥1.7 at the genus level. MALDI-TOF MS was conducted at 24th hr for the routine method for identification of microorganisms. The rapid identification results were compared with those obtained at 24 hr. Gram staining, colony morphology, agglutination with anti-Brucella serum, and biochemical tests were used for the routine identification of Brucella spp. For all microorganisms, when genus-level identification was not attained, the comparison was based on the Gram staining results. The plates were examined for the presence of polymicrobial growth at 24th hr.
Results
During the study period, a total of 243 positive BCs were analyzed. Overall, a single microorganism grew in 230 BC bottles with a species distribution of 60.0% Gram-positive cocci (GPC), 31.7% Gram-
negative bacilli (GNB), 7.4% Gram-positive bacilli (GPB), 0.9% Gram-negative coccobacilli (GNCB). In addition, 27 pathogens were isolated from polymicrobial BCs (n=13). Two different GNBs were detected in four of the polymicrobial growths, two different GPCs in five, one GNB and one GPC in three, and two GNB and one GPC in one.
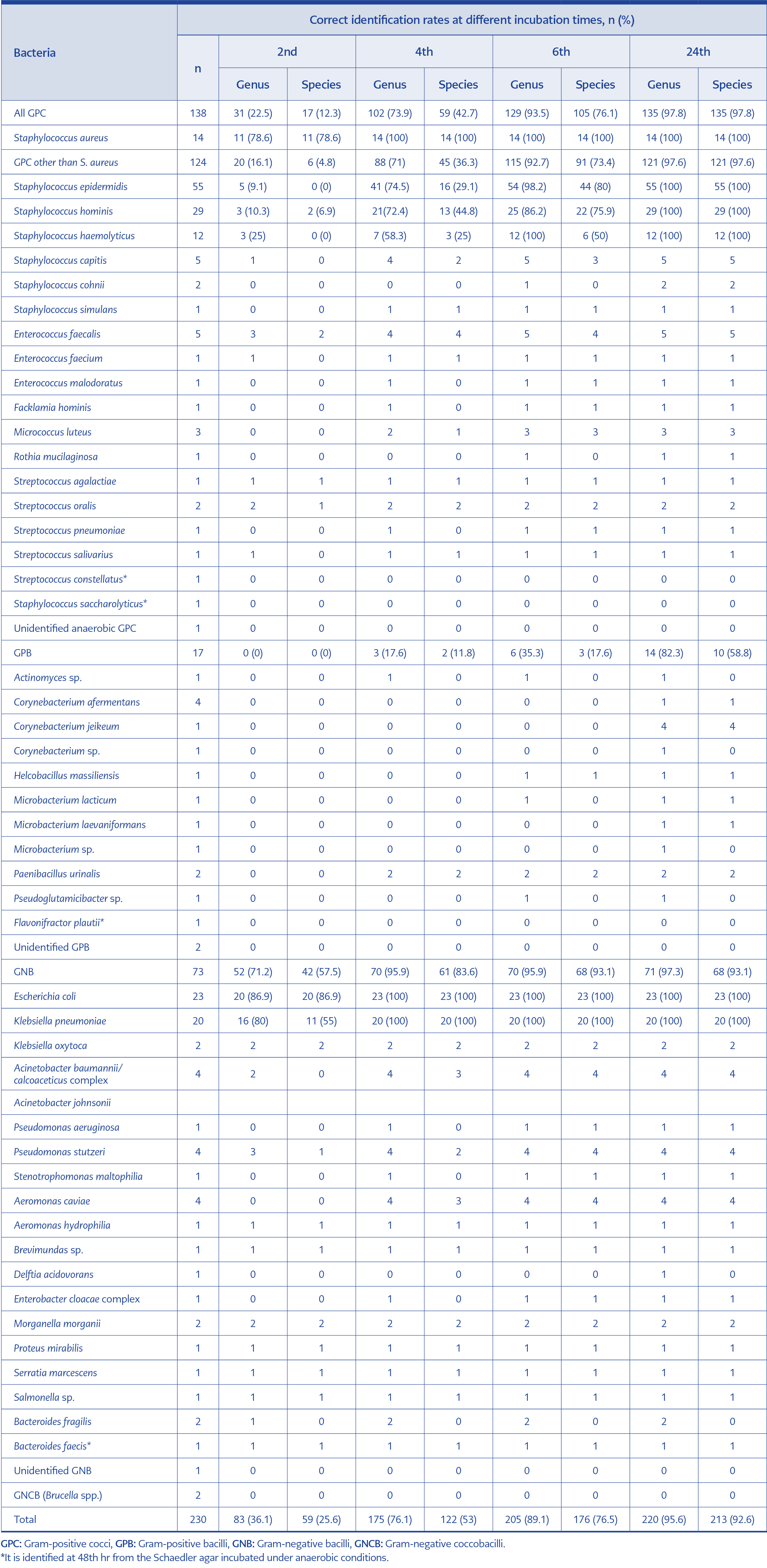
Identification rates for monomicrobial cultures are presented in Table 1. In the case of GNB, 57.5%, 83.6%, 93.1%, and 93.1% of the samples were identified at the species level at the 2nd, 4th, 6th, and 24th hours, respectively; these rates were 12.5%, 42.7%, 76.1%, 97.8% for Gram-positive cocci. All Staphylococcus aureus isolates were identified at the species level at the 4th hr. Species-level identification rates were low in Gram-positive bacilli and were 0%, 11.8%, 17.6%, and 58.8%, respectively. Species-level identification was achieved in 76.5% of all monomicrobial cultures at the 6th hour. Brucella spp. (n=2) could only be identified by conventional methods at 24 hours because it was not included in the MALDI-TOF MS database. We isolated five anaerobic bacteria, including Bacteroides fragilis, Bacteroides faecis, Staphylococcus saccharolyticus, Flavonifractor plautii and an unidentified GPC. Only the B. fragilis isolate was identified by rapid ID at 2nd hr from Columbia blood agar. Three anaerobic isolates were identified at the 48th hour from Schaedler agar incubated under anaerobic conditions. One anaerobic GPC could not be identified by the methods used in our laboratory. In polymicrobial cultures, only one of the pathogens could identified at the species level at the rate of 46.1%, 69.2%, and 84.6% at the 2nd, 4th, and 6th hours, respectively.
Discussion
The installation cost of a MALDI-TOF MS system is relatively high and often unaffordable, particularly in developing countries. However, if there is already a MALDI-TOF MS in the laboratory used for conventional (24-hour) diagnosis, the short incubation method does not require additional processes and costs. According to our work and the reports in the literature, MALDI-TOF MS, after a short period of incubation on solid media, is a simple, fast, and reliable method for the same-day identification of pathogens causing sepsis.
This study demonstrated that identification rates were higher and the time required for identification was shorter in Gram-negative bacteria and S. aureus. Identification rates were lower in other GPC compared to S. aureus and were lowest in GPB. Among the five anaerobic isolates, only one B. fragilis was identified by rapid ID. The slow growth rate of anaerobes can explain this. Previous studies reported that identification rates vary between 51.9%-96.8% and 85.8%-100% for Gram-positive and Gram-negative bacteria, respectively (6). A major reason for this is that Gram-positive bacteria have slower cell division and growth rates and form smaller biomass in a short duration. In particular, Gram-positive bacilli showed weak protein signals and identification rates were notably lower than those of other bacteria groups. The thick peptidoglycan cell walls of Gram-positive bacteria may lead to suboptimal MALDI-TOF MS spectra (7). Also, species of the bacteria, the culture media, and incubation time may affect the results. Froböse et al. reported that median time to species identification was shortest on MacConkey agar for Gram-negative rods (2.0 h; range:1.9 to 4.2 h) and on Columbia blood agar for Gram-positive cocci (4.0 h; range:1.9 to 25.0 h) (8). However, chocolate agar should be used if fastidious species are expected. Polymicrobial cultures are challenging for all rapid diagnostic methods. There was a mixed growth in 5.3% of our BCs. This highlights the importance of checking cultures at 24th hr for purity.
Our study has limitations as it used data from a single institution and was unable to compare this method with other protocols for the direct identification of microorganisms from positive BC bottles. The number of isolates included in the study was relatively low, and the interpretation of the results was limited. The constrained number of anaerobic samples represents another limitation in our study. Also, this study did not examine the clinical significance of ID from short-term cultures in determining effective antimicrobial treatment. Delport et al. reported that using a short-term incubation method by MALDI-TOF MS for identification from BCs is correlated with a decrease in the length of hospitalization and a reduction in mortality associated with bacteremia (9). Providing rapid ID results using MALDI-TOF MS without a rapid method for AST may have limited effect on directed appropriate antimicrobial therapy. However, rapid identification of the infectious agents causing bloodstream infection could be helpful for the clinician to establish an effective empirical antimicrobial therapy for patients who are in particularly critical conditions. Besides that, it is possible to combine this rapid identification method with rapid AST methods (10). Future studies should aim to combine this method with a rapid AST method and analyze whether such identification with MALDI-TOF MS leads to better clinical outcomes.
In conclusion, the application of MALDI-TOF MS following a short culture period is easy and provides same-day results. We observed a high percentage of categorical agreement with the standard identification algorithm used in our laboratory for GNB (>93.1%, 6th hr). As an outcome of our study, we have started to use the short incubation method at the 6th hour for the same-day identification of Gram-negative pathogens from positive BCs in our laboratory.