Introduction
The global increase in the rate of multi-drug resistant (MDR) bacterial infections in recent years is an important public health problem that limits the therapeutic options (1). The issue becomes particularly critical in cases of bloodstream infections (BSI) in which delay is associated with high mortality rates (2, 3). Bacteria grown in blood cultures (BC) must be subcultured on the solid media for identification (ID) and antibiotic susceptibility testing (AST) in the conventional algorithm, with species-level ID and AST results available in 48-72 hours. This may result in missing the critical time for appropriate treatment. Clinicians are faced with the choice of giving the patient narrow-spectrum antibiotic therapy and delaying effective treatment or broad-spectrum antibiotic therapy and increasing the problem of side effects and antibiotic resistance. Therefore, rapid ID and AST methods are becoming more crucial for adjustment of therapy, especially for patients with BSIs. In recent years, several biochemical, MALDI-TOF MS-based or molecular methods have been described for the rapid detection of bloodstream pathogens without subculture on the solid media (4, 5).
The European Committee on Antimicrobial Susceptibility Testing (EUCAST) developed a standardized rapid method directly from positive BCs based on EUCAST disc diffusion and validated it in a multicenter study in Europe (6, 7). Rapid antibiotic susceptibility testing (RAST) is performed directly from positive BC bottles to provide test results at the 4th, 6th, and 8th hours. EUCAST RAST method can be applied to Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii, Staphylococcus aureus, Enterococcus faecium, Enterococcus faecalis and Streptococcus pneumoniae (8). Moreover, EUCAST RAST screening recommendations are also available for the detection of extended-spectrum beta-lactamase (ESBL) and carbapenemase (9).
At our hospital, it has been a well-established practice since 1985 that almost all patients with a suspected systemic infection are consulted by the Infectious Diseases Department. These patients are evaluated by the infectious diseases consultation (IDC) team, recommendations are made for diagnosis and antimicrobial management, and they are followed on a daily basis until the infectious problem is solved. Hacettepe University Hospital has implemented a sepsis protocol for the initial diagnosis and treatment of sepsis under the guidance of the Infectious Diseases Department in 2019. The guidelines for empirical antibacterial treatment have been developed through a methodical assessment of the risk factors for antimicrobial resistance defined in the literature and local antimicrobial resistance epidemiology. In spite of these efforts, a recent study from our hospital revealed that approximately 30% of patients with bloodstream infections caused by ESBL-producing Enterobacterales received an inappropriate antibacterial treatment before antibiotic susceptibility results were reported (10). To improve the management of BSIs, a rapid ID and AST protocol was implemented at the Bacteriology Laboratory starting on October 01, 2021. This study aims to evaluate the effects of rapid ID and RAST on the clinician’s treatment decision and to determine the possible added value to patient management.
Materials and Methods
This prospective observational study analyzed BCs collected between October 01, 2021, and December 31, 2021, at the Central Bacteriology Laboratory of the Hacettepe University Hospital in Ankara, Türkiye. BC bottles (BD BACTEC™ Plus Aerobic/Lytic Anaerobic; Becton Dickinson and Company, USA) were incubated for five days or until flagged positive using the BD BACTEC™ FX (Becton Dickinson and Company, USA) instrument. After a positive signal, direct microscopic examination by Gram staining was performed immediately. Positive BC samples were inoculated to sheep blood agar (RTA Labs, TÜrkiye), eosin methylene blue agar (RTA Labs, Türkiye), and chocolate agar (Becton Dickinson and Company, USA) for aerobic culture. In addition, Schaedler agar (Becton Dickinson and Company, USA) was used and incubated in anaerobic conditions for anaerobic BC bottles.
For rapid ID, cell pellets obtained after centrifugation with serum separator gel tubes and MALDI-TOF-MS (Biotyper IVD 4.2.80; Bruker Daltonics, GmbH, Germany) were used. This method was described earlier by Wu et al. (11). A log (score) of ≥2.0 was considered an accurate species-level identification. If the log (score) was ≥1.7 and <2.0, genus-level identification was accepted. Results with a log (score) <1.70 were considered unacceptable. RAST was performed using Oxoid antibiotic discs (Thermo Fisher Scientific, USA) on Mueller Hinton agar (Becton Dickinson and Company, USA) according to the recommendations of EUCAST (v1.1, 2019) (12). Zone diameters were interpreted using “Breakpoints for Rapid AST” directly from positive blood culture bottles (v3.0, 2021) as follows: not readable (poor growth); S (susceptible); I (susceptible, increased exposure); R (resistant); and ATU (area of technical uncertainty), which denotes a weak distinction between susceptibility groups (8). Since no categorization was done, results in the ATU were excluded from error calculations. Three categories were used to describe categorical errors: very major error (VME; RAST=S and reference=R), major error (ME; RAST=R and reference=S), and minor error (mE; RAST=S or R and reference=I or RAST=I and reference=S or R). In conventional AST, the BD Phoenix™ M50 instrument (Becton Dickinson and Company, USA) was used for E., K. pneumoniae, P. aeruginosa, S. aureus, and E. faecalis/faecium strains. Carbapenem/vancomycin resistance or any unexpected resistant/susceptible phenotype by BD Phoenix™ M50 was confirmed by E-test (bioMérieux, France). The disk diffusion method was applied to A. baumannii isolates. Minimum inhibitory concentration (MIC) values and zone diameters were interpreted according to EUCAST Breakpoint Tables (version 11.0, 2021) (13).
RAST, as a part of daily laboratory care, was planned by the Antimicrobial Stewardship Committee as a quality improvement initiative. At baseline, senior consultants and registrars in the Infectious Diseases and Clinical Microbiology Department were informed that rapid ID results would be reported by the bacteriology laboratory if a BC was flagged as positive. It was also announced that the RAST results would be reported on the same day for E. coli, K. pneumoniae, P. aeruginosa, A. baumannii, S. aureus, and E. faecalis/faecium strains, for which the standards for RAST have been established by EUCAST. A warning note was added to all reports that the results of rapid ID and RAST were preliminary and that the results of the standard method should be followed. The impact of these reports on the clinician’s treatment decision was assessed through clinical documentation. The appropriateness of definitive antimicrobial therapy was assessed by an infectious diseases specialist based on institutional antimicrobial prescribing guidelines, AST results and clinical data. The results of using rapid ID and RAST as a new approach were presented in a report to the hospital administration. The Hacettepe University Observational Research Ethics Committee approved the study on February 21, 2023, with the decision number 23/133.
Results
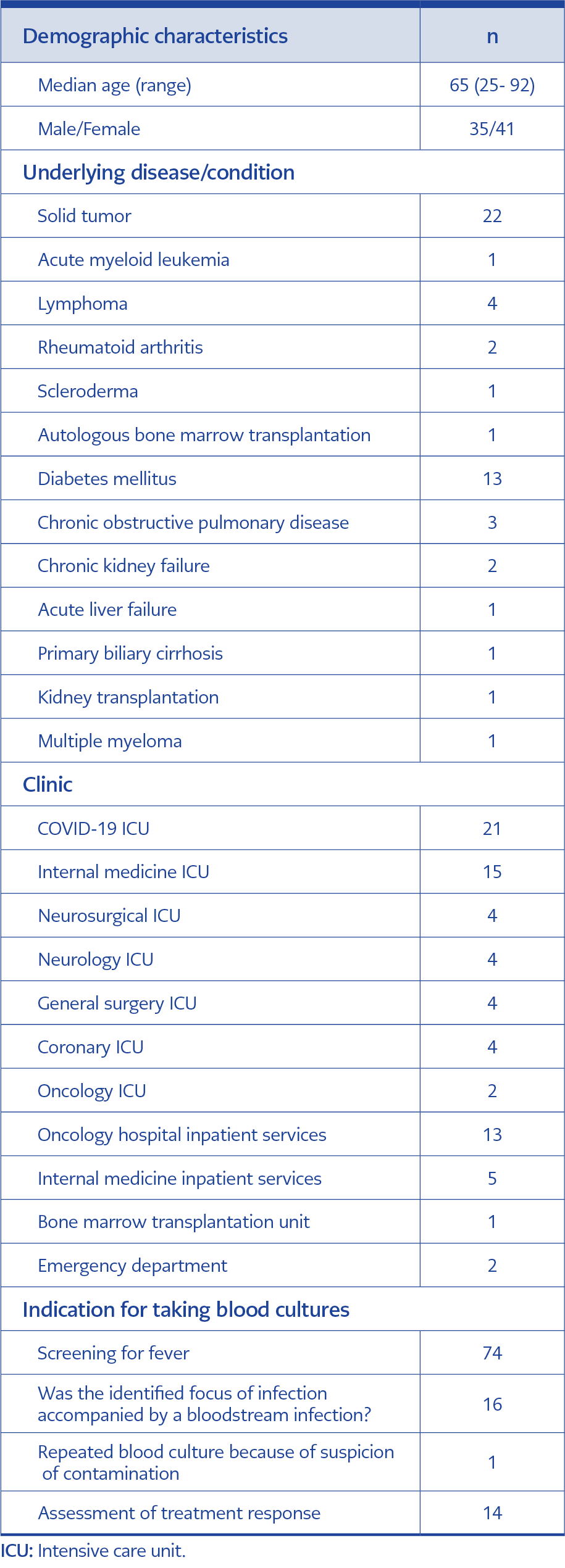
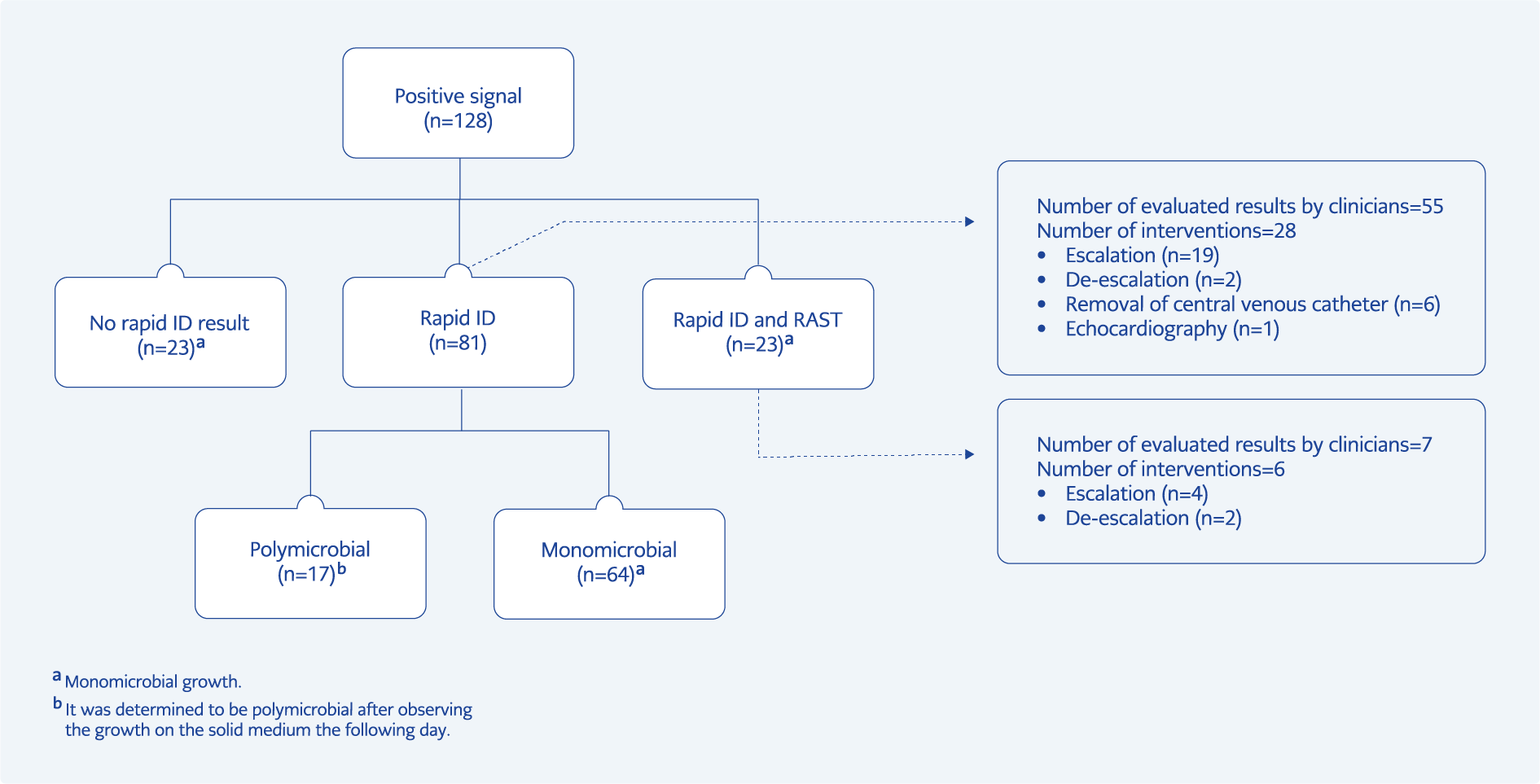
In total, 128 BC bottles from 86 patients were processed. Identification was achieved in 105 (82.1%) bottles taken from 76 patients with the rapid ID method used in the study. Of the 76 patients, 35 were men and 41 were women. The median age was 65 years (range: 25-92 years). Fifty-six (73.7%) patients were in intensive care units, and 21 (27.6%) were treated with a diagnosis of COVID-19 disease. Forty-nine (64.5%) patients had at least one underlying disease that could cause immunosuppression. The most common underlying diseases were solid tumors and diabetes mellitus. The most common indication for obtaining BCs was evaluation for fever (Table 1). The patients’ clinical records showed that the rapid identification results were evaluated by the IDC team on the same day for 55 (52.4%) of 105 BCs. Based on the evaluation of the rapid ID results, new treatments or interventions were ordered for 28 (26.7%) of 105 cultures, including escalation (n=19), de-escalation (n=2), central venous catheter removal (n=6), and echocardiography (n=1). Treatment remained unchanged for ten cultures, and the results of 17 cultures were interpreted as skin microbiota contamination (Figure 1). If the rapid identification results had been evaluated on the same day, additional recommendations to the current treatment could have been made for eight (7.6%) more cultures, including escalation (n=3), de-escalation (n=4), and removal of the central venous catheter (n=1).
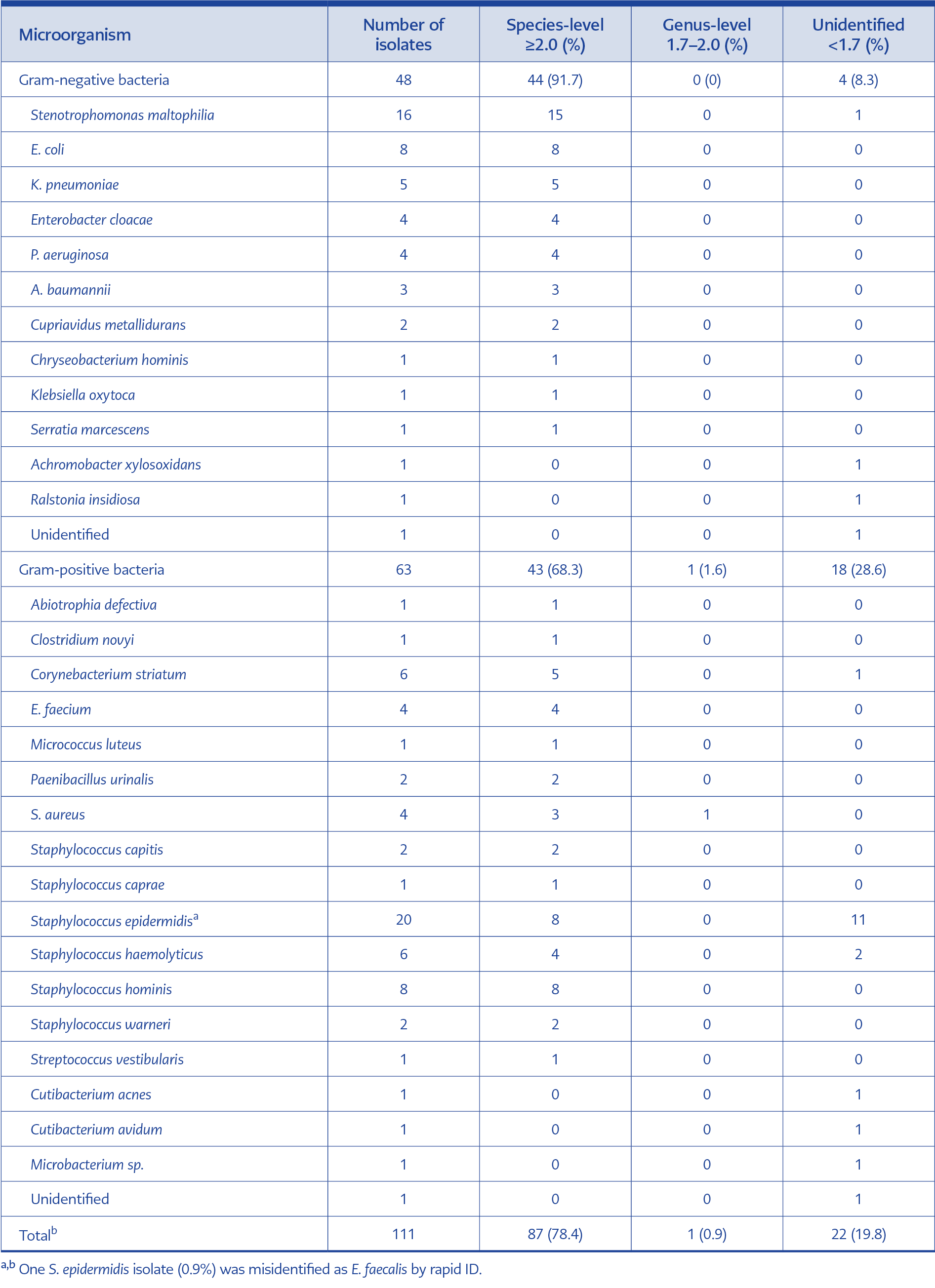
Table 2. Comparison of rapid identification results with MALDI-TOF MS results for monomicrobial cultures (n=110)
Gram stain showed monomicrobial infection in all BCs processed for rapid ID and RAST, but 17 were found to be polymicrobial after 24 hours of incubation on solid media (Figure 1). In monomicrobial BCs (n=111), 48 (43.2%) were Gram-negative, and 63 (56.8) were Gram-positive (Table 2). Overall, 79.3% and 78.4% of these bacteria were directly identified at the genus and species level, respectively. There was a difference in the agreement of rapid ID and conventional ID between Gram-positive and Gram-negative bacteria: 68.3% of Gram-positive bacteria were correctly identified, compared to 91.7% of Gram-negative bacteria at the species level. One of the S. epidermidis isolates (0.9%) was misidentified as E. faecalis by rapid ID. In polymicrobial BCs, one of the growing bacteria was identified to the genus level in all bottles; species-level identification was achieved in 15 BC (88.2%) bottles (Table 3). A total of the 42 bacteria were members of the skin microbiota, and 40 of them were accepted as BC contamination.

Table 3. Comparison of rapid identification results and MALDI-TOF MS identification results at 24th hours in polymicrobial cultures (n=17)
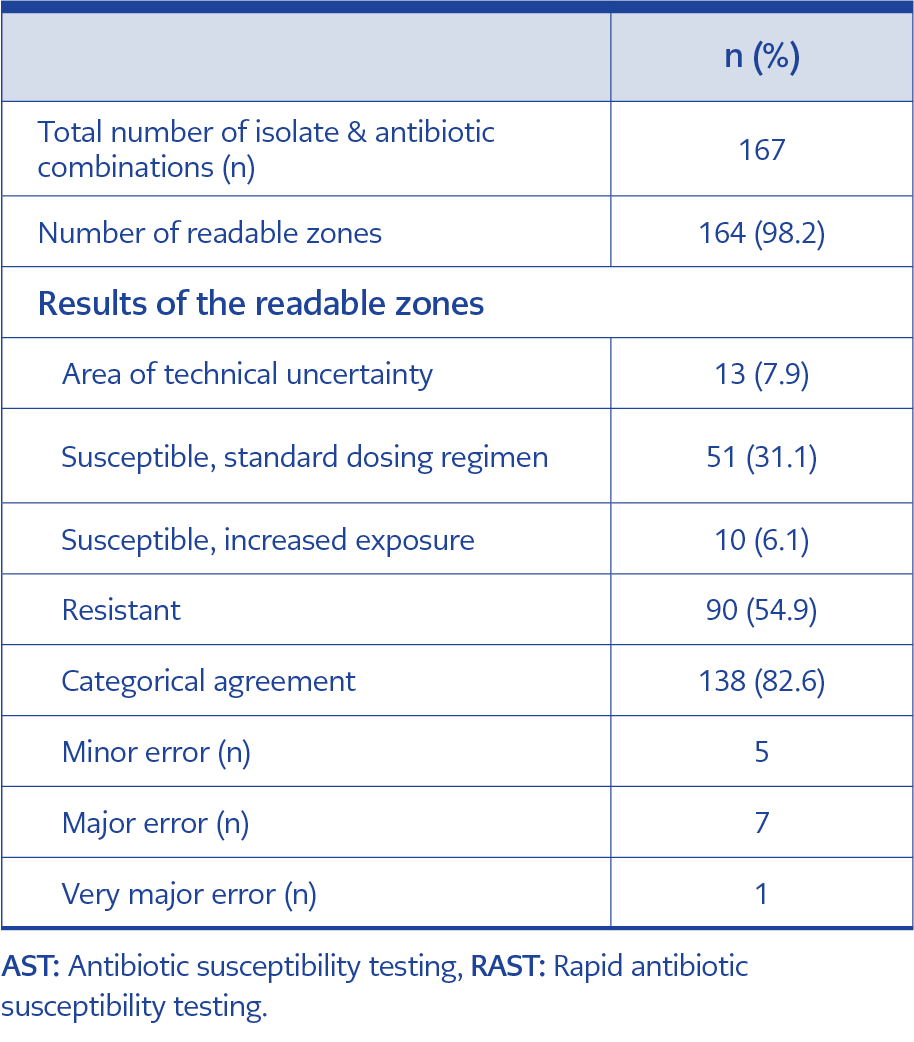
Following rapid ID, RAST was performed on non-duplicate 24 isolates from monomicrobial BCs (Table 4). Three-zone diameters (E. coli-trimethoprim/sulfamethoxazole, E. coli-amikacin, P. aeruginosa-ciprofloxacin) were not readable. The categorical agreement of RAST with conventional AST was 82.6% at the 6th hour for all isolates. The agreement between conventional AST was 84.5% for Gram-negative bacteria (n=17) and 72.0% for Gram-positive bacteria (n=7). Of all isolate-antibiotic combinations, 54.9% were found to be resistant. ATU was observed for 13 isolate-antibiotic combinations including E. faecium-vancomycin (n=4), E. coli-piperacillin/tazobactam (n=3), E. coli-amikacin (n=2), E. coli-gentamicin (n=2), E. coli-trimethoprim/sulfamethoxazole (n=1), K. pneumoniae-piperacillin/tazobactam (n=1). One VME was detected for clindamycin in S. aureus. Seven MEs were determined for E. coli-amikacin (n=2), E. coli-gentamicin (n=1), E. coli-piperacillin/tazobactam (n=1), E. coli-ciprofloxacin (n=1), S. aureus-clindamycin (n=1), E. faecium-high level gentamicin resistance (n=1). Five minor errors were observed for E. coli-ceftazidime (n=2), K. pneumoniae-ceftazidime (n=1), P. aeruginosa-ceftazidime (n=1), P. aeruginosa– piperacillin/tazobactam (n=1). The IDC team evaluated the results of the antimicrobial susceptibility profile of the pathogen for seven BCs on the same day. Treatment was escalated in four cultures, de-escalated in two cultures and remained unchanged in one culture. If the RAST results had been evaluated on the same day, four more cultures could have been de-escalated, and seven more escalated according to the susceptibility results.
Discussion
Rapid ID and AST of bacterial isolates in blood cultures can lead to earlier modifications in antimicrobial treatment in patients with BSIs, but data with an emphasis on how clinicians assess rapid ID and AST results and their implications for treatment are sparse in the literature. In our study, early therapy modifications were made for 36.8% of patients as a consequence of the same-day examination of rapid ID results. Rapid ID results had not been assessed on the same day in 27.6% of the patients. If they had been assessed, there was a potential to modify the treatment in 10.5% of the patients. This finding implies that it is crucial to facilitate close communication between the IDC and the laboratory teams.
A similar study by Strubbe et al. reported that AST-guided intervention could be made earlier in 44.5% of the patients with the implementation of the EUCAST RAST method. The authors noted that AST reporting was effective in correcting ineffective empirical therapy in 6.3% of the patients (14). This latter study differs from ours in several aspects: first, it was carried out in a setting characterized by a low prevalence of MDR; second, RAST results were not provided to the clinician; and finally, the appropriateness of definitive antimicrobial therapy was evaluated by a single microbiologist. Valentin et al. examined the clinical implications inferred by infectious diseases physicians based on RAST results during same-day bedside consultations (15). IDC was conducted for 134 patients following the acquisition of RAST results. Changes or initiation of antimicrobial treatments were made in 73 (54.5%) patients, and 84 additional measures (imaging studies, surgery, and additional resistance testing) were recommended for 62 (46.3%) patients. Tayşi et al. determined the effects of p-RAST (a variation of the EUCAST RAST method) results on the treatment decision of the clinicians (16). In their study, p-RAST results were reported to the infectious diseases physician by direct calls. The study showed that escalation of the antibacterial regimen was made in 31.1% (45/145) of the patients, and de-escalation could be made in 22.1% (32/145) of the patients. French et al. showed that faster ID of bacterial isolates from BCs using MALDI-TOF MS has a direct clinical effect and facilitates the prompt selection of an appropriate antimicrobial agent, even in the absence of susceptibility testing (17).
Advanced molecular identification methods are available for the rapid identification of bloodstream pathogens. Nonetheless, their integration into routine laboratory workflow is challenging due to the high cost (18). In recent years, MALDI TOF MS has emerged as a fast and reliable technique for microbial diagnosis. The identification of microorganisms from a positive BC bottle can be achieved directly through the MALDI-TOF MS system, utilizing either “in-house” methods or commercial kits along with additional software modules like the MBT-Sepsityper (Bruker Daltonics, GmbH, Germany).
The major disadvantage of commercial kits is their high cost (5, 18). Using serum separator gel tubes to obtain bacterial pellets and perform ID with MALDI-TOF MS is a cheap and rapid in-house method. In previous studies with serum separator tubes, rates of correct identification for Gram-negative bacteria varied from 93% to 100% and 32% to 92% for Gram-positive bacteria (11, 19-21). It should be emphasized that lower cut-off levels were used for identification in some studies, potentially yielding higher identification rates compared to those with traditional cut-off levels (≥2.0, species-level; <2.0 and ≥1.7, genus-level; ≤1.7, unidentified). Our species-level identification rates with serum separator tubes were 91.7% in Gram-negative and 69.4% in Gram-positive bacteria, using classical cut-off levels. The RAST results in our study showed a lower degree of agreement with routine susceptibility testing methodology compared to the previous reports (16, 22, 23). The most likely reason for this is our small sample size. Additionally, the experience of the personnel performing RAST, different brands used, and variations in the routine AST method may explain the differences in the studies.
In this study, 17 (13.3%) of the cultures were identified as polymicrobial the following day by observation of the growth on solid media. While some polymicrobial cultures can be identified by Gram staining, some cultures may be misinterpreted as monomicrobial if the isolates have similar Gram staining characteristics and morphology. This poses several challenges in the diagnosis and treatment of infections including the identification of causative bacteria, interference with AST, difficulty in choosing the appropriate antibiotic, and delayed treatment. It is important to check growth on solid media the following day to obtain final results when using the rapid ID method to process bacteria (16).
Another important finding of our study was that 40 of 105 (38.1%) isolates were regarded as skin microbiota contamination. This indicates that the proper practice of obtaining BCs has to be reinforced at our center. We believe that the most likely reason for a high contamination rate is that BCs are not taken by a specific team. Therefore, the inexperienced intern or physician might apply the wrong skin disinfection or blood drawing technique, and effective feedback cannot be given because there is no record of who obtained the BC. Blood collection by trained phlebotomy personnel and education on proper techniques may prevent this problem. In addition, using the BC contamination rate as a quality score for the physician who requested the culture and the associated clinic could potentially address the issue.
This study has several limitations. First, its sample size is small and conducted in a single center. Second, RAST readings were not performed and compared at different times due to the low number of personnel and laboratory operating hours. Third, although we demonstrated that rapid ID and AST reporting influenced the revision of empirical therapy, we did not assess its impact on patients. As of our current knowledge, the effectiveness of these early interventions alone in improving clinical outcomes is uncertain. Previous research has demonstrated the clinical benefits of shortened time to rapid ID and AST, including a higher proportion of optimally treated bloodstream infections, decreased rate of transfer to ICU, decreased length of stay and reduced mortality (24-26). In contrast, the investigators of the RAPIDO trial found that despite providing the results to clinicians sooner, the rapid ID of bloodstream pathogens by MALDI-TOF MS did not lower patient mortality (27). In a study by Anton-Vazquez et al., the median time to optimal antibiotic selection based on AST results was significantly shorter compared to the conventional group (28). Nevertheless, no significant differences were noted in terms of 28-day mortality or length of stay. Similarly, Berinson et al. found that although optimal therapy was initiated one day earlier in the RAST group (n=54) compared to the historical group (n=54), the difference in 30-day mortality rates was not statistically significant (29).
In this study, we investigated how clinicians evaluated the rapid ID and AST results and their impact on patient treatment. Our findings support the idea that rapid ID and AST have the potential to make a significant contribution to guiding antibiotic therapy. Further larger-scale research is needed to determine the effect of reporting rapid results on patient outcomes. In addition, our study underlined the importance of close and timely communication between clinicians and the laboratory while performing rapid ID and AST.