Introduction
SARS-CoV-2 has affected the whole world while the number of cases is increasing rapidly. Several reinfection cases have been reported with the rise in the number of infected people (1). It is important to study these reinfection cases as the knowledge gained can be used in critical decisions to control the disease and plan the treatment options for the patients. It is also expected that the information about this subject can shed light on vaccine development studies (1,2).
Centers for Diseases Control and Prevention (CDC) defines reinfection as the diagnosis of COVID-19 after the patient has recovered from the first infection and tested negative by PCR for more than 90 days. The confirmation for reinfection includes a verified first episode of infection, proof of reinfection with RT-PCR results below 35 Ct value and negative RT-PCR results between the two episodes of infection (1,2). In addition, viral sequence analysis must also be performed to confirm the case as reinfection. According to the European Centre for Disease Prevention and Control (ECDC), countries that have reported reinfection cases so far are the USA, Israel, Ecuador, Hong Kong, India and Belgium (3).
Our center is a university hospital that has been actively dealing with COVID-19 patients since March 17, 2020, the date when the first COVID-19 case was confirmed in our hospital. From March 2020 to December 2020, approximately 4000 COVID-19 patients had been monitored in our center. During that period, two patients suspected of reinfection based on their clinical symptoms and laboratory results were confirmed as reinfection cases after performing viral sequence analysis. In this article, we discussed the clinical, laboratory and genomic analysis of these two cases.
Clinical and Laboratory Findings of Cases
Case 1
First infection
The patient was a 31-year-old female infectious disease physician who had been actively following COVID-19 patients. She was confirmed positive for SARS-CoV-2 upon RT-PCR testing of combined nasopharyngeal/oropharyngeal swap (NOP) sample collected on April 11, 2020. The RT-PCR testing was performed using the Bio-speedy® SARS-CoV-2 Double Gene RT-qPCR kit (Bioeksen, Turkey), targeting ORF1ab and N regions. The patient had shown symptoms of fever and myalgia, which had started two days before the sample collection. Blood count parameters results were determined as follows: Leukocyte count: 10,890/L (4,500-11,000), lymphocyte count: 370/L (1,500-4,000), neutrophil count: 10,170 /L (1,800-7,700). All biomarkers like renal functions and livers enzymes were normal. Chest computerized tomography (CT) findings were unremarkable.
The patient was breastfeeding, and she was treated with hydroxychloroquine 2x200mg and subcutaneous low-molecular-weight heparin (LMWH) 0.4 1×1 subcutaneous. The total treatment period was five days.
The patient’s complaints were resolved within two days, and no complications occurred in the follow-up. After resolving the complaints, two consecutive nasopharyngeal samples were taken on April 19, 2020, and April 22, 2020, for RT-PCR analysis, and both revealed negative for SARS-CoV-2 RNA. She was also tested negative for the presence of anti-SARS-CoV-2 total anti-nucleocapsid antibody (Elecsys® Anti-SARS-CoV-2, Roche Diagnostics, Germany) on June 17, 2020.
Reinfection
Eighteen weeks after primary infection, the patient exhibited typical symptoms of COVID-19, including sore throat and myalgia with the feeling of pressure on the chest for the last two days. RT-PCR analysis of combined NOP was again positive for SARS-CoV-2 RNA. In the CT, ground-glass areas were seen in the left middle lobe of the lung. All blood parameters were within normal limits. No viruses other than SARS-CoV-2 was detected in the simultaneously taken respiratory viral multiplex PCR panel (FTD Respiratory Pathogens 21, Fast Track Diagnostics, Luxembourg).
During that period, favipiravir treatment was offered, but as she refused to use it, she was followed on LMWH. The patient’s complaints were resolved within three days. On January 15, 2021, the antibody test result was positive for anti-SARS-COV-2 total nucleocapsid antibodies (Elecsys® Anti-SARS-CoV-2, Roche Diagnostics, Germany) (COI=115.8).
Case2
First infection
The second case was a 46-year-old male working as a laboratory technician. He had a dry cough that started two days before the RT-PCR test. The SARS-CoV-2 RT-PCR test performed on April 2, 2020, was found to be positive. Except for ALT (68 U/L) and GGT (95 U/L), all other blood biomarkers were within normal limits. Chest CT was unremarkable. The patient received hydroxychloroquine 2x200mg orally and LMWH for five days. His complaints subsided after the third day, and no symptoms were seen in the follow-up. Two consecutive NOP swap samples taken on April 14, 2020, and April 16, 2020, were SARS-CoV-2 negative by RT-PCR testing.
The SARS-CoV-2 antibody test results of the patient taken on June 25, 2021, and July 14, 2021, were negative for anti-SARS-COV-2 total nucleocapsid antibodies (Elecsys® Anti-SARS-CoV-2, Roche Diagnostics, Germany).
Reinfection
After five months from the first infection, he developed fever, myalgia and fatigue on September 2, 2020. The NOP swap sample taken for SARS-CoV-2 RT-PCR was positive again. Because he had no cough on admission, a radiological examination was not performed. All biomarkers in the patient’s blood parameters were normal. The patient was treated with favipiravir and LMWH. After 48 hours of therapy, his complaints were resolved, and no complications were seen in follow-up.
Sequence Analysis of the SARS-CoV-2 Strains
For genomic sequence analysis, total nucleic acid, including the SARS-CoV-2 RNA, was extracted from the NOP samples of the patients obtained during the first and second infections using the EZ1 Virus Mini Kit v2.0 (Qiagen, USA). Isolated RNA was then processed for sequencing using the PARAGON CleanPlex® SARS-CoV-2 Panel kit. The RNA samples were first transcribed into cDNA, and multiplex PCR was performed according to the kit manual. Following digestion reaction and second PCR reaction using index primers, a sample pool was prepared before sequencing. For sample pool preparation, each PCR product was first diluted to 1 nM then to 100 pM. Ten μl of each of the 100 pM samples was pooled together to make the 100 pM sample pool. This sample pool was then sequenced using the Illumina iSeq100. After sequencing, FASTQ data was analyzed for the quality and presence of mutations using the Genome Detective platform. In addition, the Nextstrain platform was used to control the quality of the sequences and assign them to their respective clades. In addition, lineage classification of the isolates was done using the https://pangolin.cog-uk.io/. FASTA data obtained from Genome Detective was transferred to the Molecular Evolutionary Genetics Analysis (MEGA) software package program to align and construct the phylogenetic tree. BLAST and GISAID searches were also performed to detect records with high identity scores to our sequences. Phylogenetic comparison between the sequences was performed in the MEGA software using distance matrix/ UPGMA with p-distance parameter and 1000 bootstrap value.
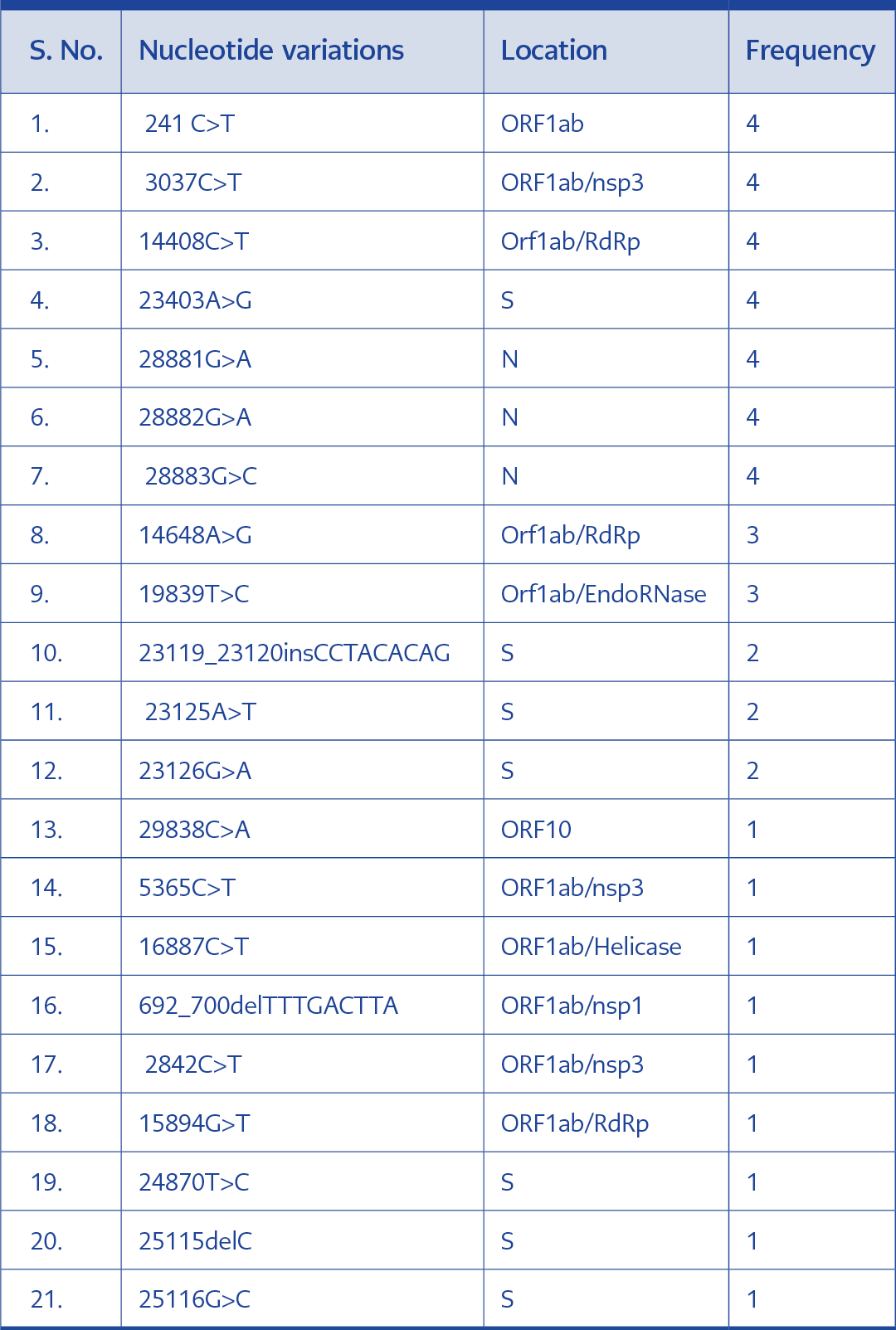
Upon the sequence analysis, a total of 20 nucleotide variations were confirmed in the four samples. Seven of them were seen in all four sequences. These are 241C>T, 3037C>T, 14408C>T, 23403A>G, 28881G>A, 28882G>A and 28883G>C. All four isolates were members of the 20B clade and the B.1.1 lineage of SARS-CoV-2 (4-8)(Table 1).
Case 1
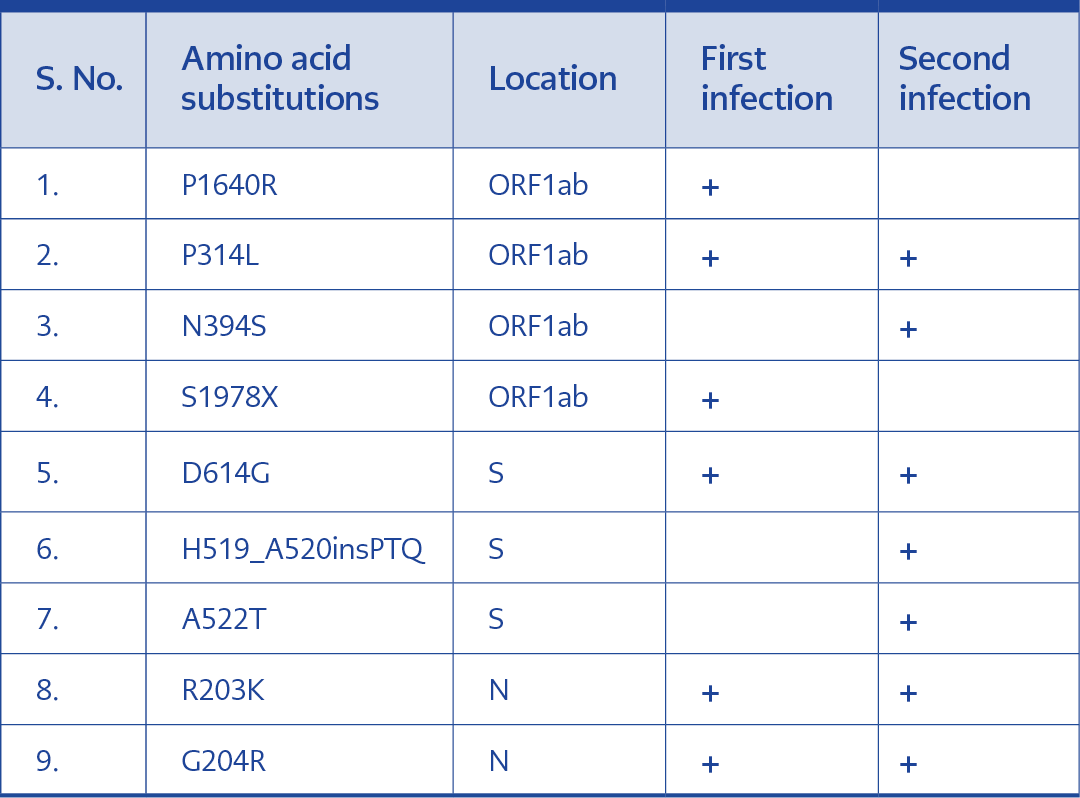
The specimens collected from this patient exhibited different nucleotide variations. Variations 241C>T, 3037C>T, 14408C>T, 14648A>G, 19839T>C, 23403A>G, 28881G>A, 28882G>A, 28883G>C, 29838C>A were seen in in the sample collected during the first infection. These variations were also reported in the samples taken during reinfection. In this case, nucleotide variations exclusively recorded in the reinfection sample were 5365C>T, 16887C>T, 23125A>T, 23126G>A and 23119_23120insCCTACACAG. A total of four amino acid substitutions were observed in the ORF1ab region of the two samples. Two of these substitutions (P314L and N394S) were seen in both samples. While the substitution S1978 was only seen in the first infection, N394S was exclusive to the second infection. Interestingly two other additional amino acid insertions and substitutions (H519_A520insPTQ and A522T)(Table 2).
Case 2
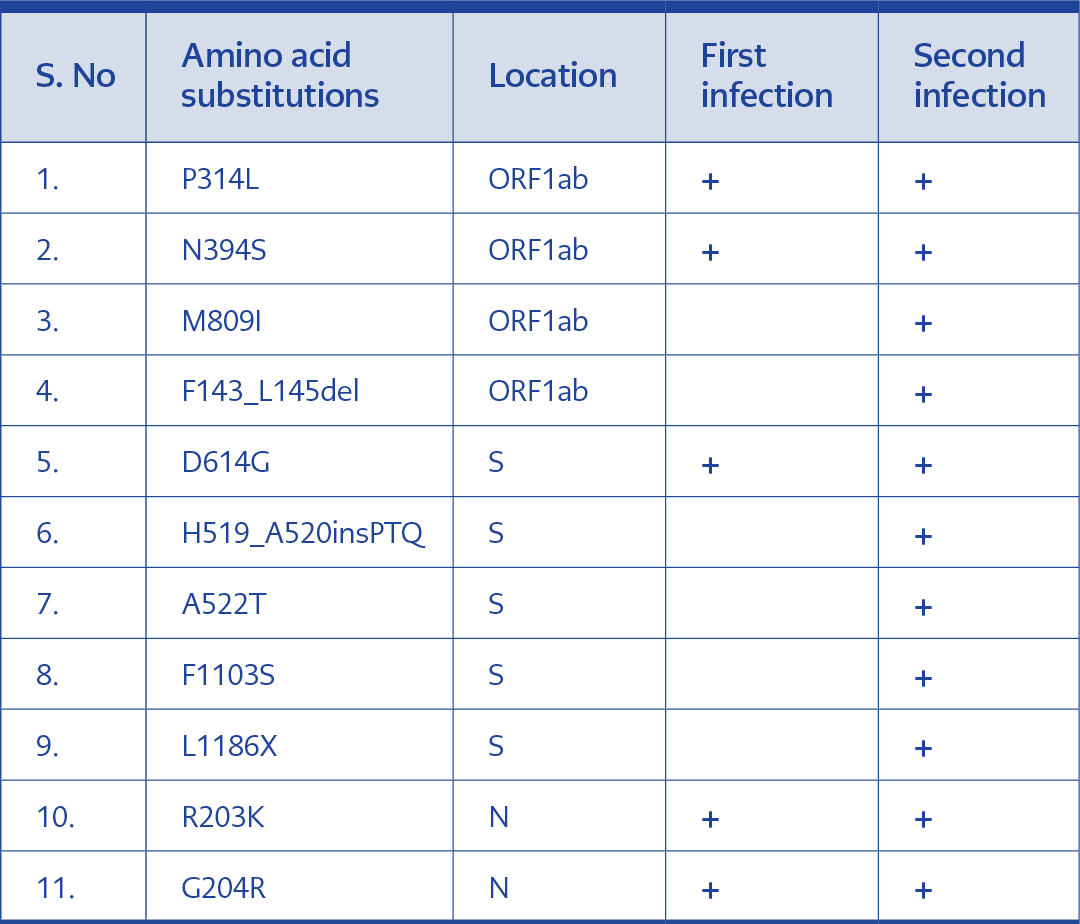
Sequencing results of the samples collected from this patient during the first infection showed variations at locations 241C>T, 3037C>T, 14408C>T, 14648A>G, 19839T>C, 23403A>G, 24453A>G, 28881G>A, 28882G>A and 28883G>C. Similar to the previous case, all these variations except 24453A>G were also seen in the reinfection specimens. Moreover, mutations 692_700delTTTGACTTA, 2842C>T, 15894G>T, 23119_23120insCCTACACAG, 23125A>T, 23126G>A, 24870T>C, 25115delC, 25116G>C were specifically identified in the reinfection samples. Sequencing results from this patient has also exhibited a higher number of amino acid substitutions during the second infection. Amino acid substitutions M809I in the ORF1ab, A522T, F1103S, L1186X and insertions H519_A520insPTQ in the S protein were among the mutations exclusively seen in the second infection (Table 3).
Discussion
Since the first COVID-19 case in December 2019, new information about the pathology and genomic arrangement and mutations of the virus has continued to emerge. Identifying the mutations that may occur in the coding regions of the genome may affect the severity of the disease as well as the results of antibody testing (4,9).
The first case of reinfection was reported in Hong Kong in August 2020, and since then, several similar cases were published. As of January 2021, cases of COVID-19 reinfection were reported in Hong Kong, The Netherlands, Belgium, Ecuador, Israel, India, Australia and the United States (10-14).
As of March 2021, 4700 COVID-19 patients were followed in our center, and it was thought that 11 of them might have clinical reinfection. However, out of the 11 patients, only two cases of reinfection were confirmed by PCR testing and performing sequence analysis.
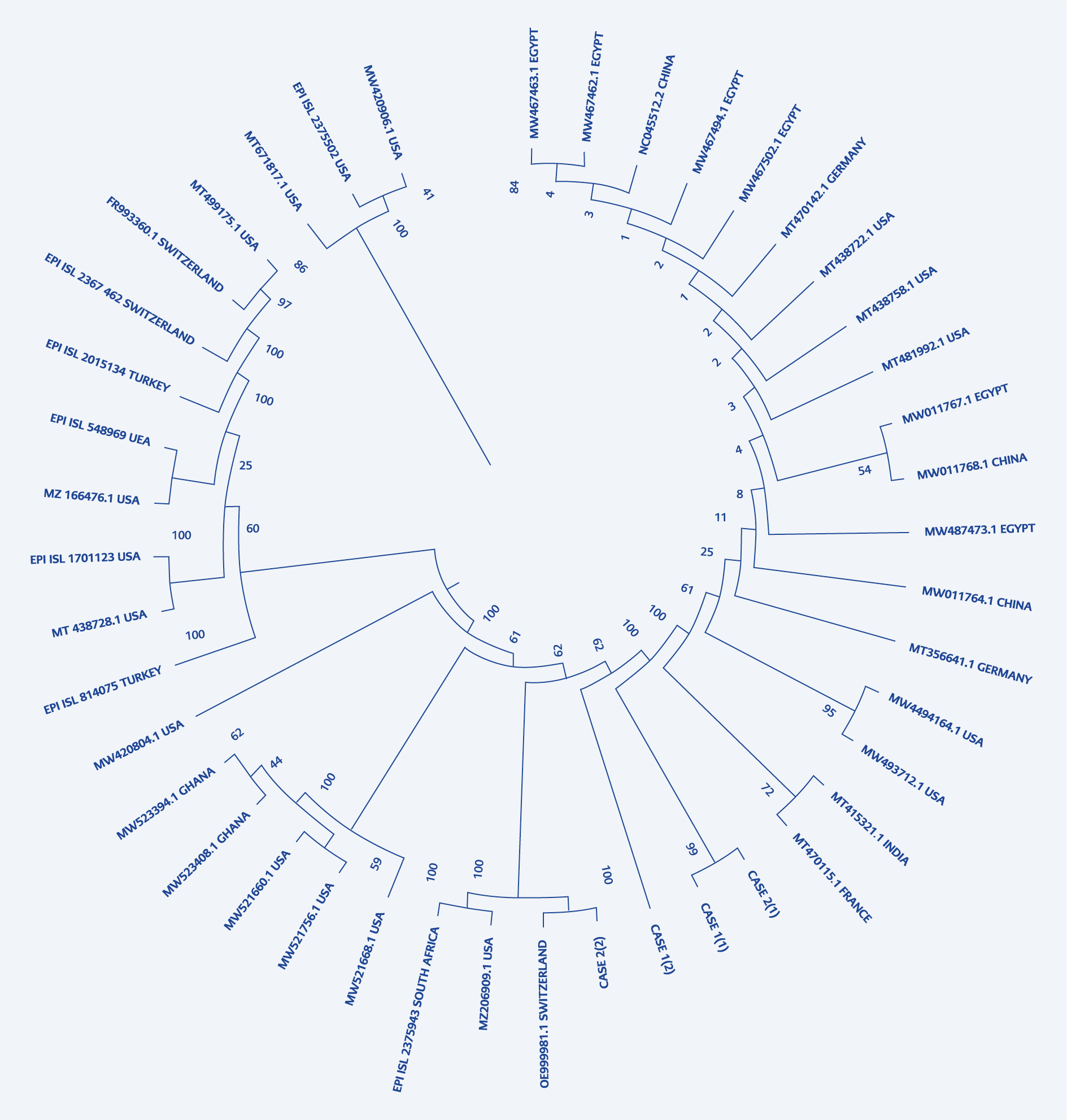
The sequence analysis of the cases revealed the different nucleotide variations in the reinfection compared with the previous ones. Even though the sequences seemed to appear of the same clade, i.e. 20B, significant nucleotide variations and amino acid substitutions were seen. Upon BLAST and GISAID searches, the sequences from the previous infections in these two cases matched with isolates from France and India. These sequences were recorded in mid-May-June and September 2020. Interestingly these two isolates from the first infections belonged to the same node in the phylogenetic tree. The reinfection isolate from the second case had recorded high homology with OE999981.1 from Switzerland, which was documented in January 2021 (Figure 1).
Reinfections cases in the literature show us reinfections might be milder or severe compared to the first infection. However, the reasons for these different situations are not known well (10-14).
Amino acid mutation D614G in the S protein along with P314L in the RdRp of the ORF was present in all the sequences. In comparison with other studies, these mutations seem to be the main ones circulating in the community in Turkey (4). These co-occurring mutations have recently been described as one of the major SARS-CoV-2 variants in Europe (5). It has been reported that the presence of the D614G mutation can increase the viral load of the virus (6). In addition, this mutation augments virus infection by increasing the total S protein incorporated into the virion (7). The amino acid insertions H519_A520insPTQ and deletion F143_L145del were not unique to the reinfections but were also present in our other sequences that have not been published yet. So, the effect of these mutations on the transmission and pathogenicity of the virus must be assessed.
The majority of the patients represented in the literature are health care workers. Similarly, the two patients described in this article are also health care workers who are more likely to be exposed to different strains of the virus. Easier accessibility of hospitals and diagnostic facilities and increased awareness of symptoms of the infection can also be reasons why COVID-19 reinfections are mainly reported in health care workers. The patients in our study did not show significant variance in the severity of the signs and symptoms between the infection episodes. Similar patterns of the severity of the disease were also reported in Hong Kong (8). Even though it is not fully confirmed, the severity could result because of increased viral load, virulence in the second infection (9).
It is well known that coronaviruses cause reinfections, similar to other upper respiratory tract infecting viruses. A human being’s long-term response to these viruses may not be enough to protect from a second episode of infection by the virus. For instance, previous studies on MERS and SARS-CoV infections have shown that total binding and neutralizing antibodies decrease slowly over time (10). In this study, the antibody tests for these cases in the first episode of the infection were negative, but a significant positive result was reported in the second. The same findings were reported in Hong Kong and Ecuador (15,16). Some studies reported that the amount of antibodies was proportional to the severity of the disease, and hence mild cases of infection might show decreased long-lasting antibody production (17). However, further study is required to assess whether the second infection’s severity results from changes in the immunocompetency of the patients or due to the presence of various new mutations in the virus.
Conclusion
Defining reinfections microbiologically and increasing knowledge on this field is very important in the prevention and a better understanding of the pathogenesis of the virus. In addition to this, further studies of reinfection cases can help us understand the response of the immune system. On the other hand, it is crucial to assess the efficacy of the currently used vaccines against the virus. Therefore, further studies involving re-infections and immune responses exerted by the vaccines must be conducted.