Introduction
Even though Candida albicans is the most isolated fungus from clinical specimens, in recent years, there has been an increase in isolation of non-Candida spp. including Trichosporon asahii. T. asahii is an arthroconidial yeast belonging to the Trichosporonaceae family of the Basidiomycota phylum taxonomically. It can be found in nature as well as in human microbiota of skin, oral cavity, vagina and respiratory and gastrointestinal systems. This fungus can cause superficial and deep-seated invasive infections (5, 11, 18, 20). Direct microscopic examination of clinical materials, morphological and microscopic examination of the colonies produced in culture and assimilation tests are important for the preliminary identification of the fungus, but mass spectrometry (MS) or molecular methods should be performed for correct identification. For the molecular identification of T. asahii, 26S rDNA internal transcribed spacer (ITS), D1/D2 and intergenic spacer (IGS) regions are used (10, 18). T. asahii may exhibit different genotypes in different geographic regions. Fifteen genotypes of T. asahii have been found so far (9). In several in vitro studies, high minimal inhibitory concentration (MIC) values for amphotericin B (AMB) and fluconazole (FCZ) have been reported (11). It has also been reported that new azoles as posaconazole (POS), voriconazole (VOR) and ravuconazole are more active than FCZ and AMB on this fungus (21).
We aimed to determine genotypic variations and antifungal susceptibilities of 70 T. asahii clinical isolates and describe epidemiological data based on the possible relationship between the two parameters.
Materials and Methods
Microorganisms
A total of 70 T. asahii strains regardless of their clinical importance, isolated from the clinical samples of 70 different patients (61 hospitalized, 9 outpatient) admitted to Marmara University Pendik Training and Research Hospital, between the years 2006-2016, with the exception of 2011-2012 and only one strain in January 2017, were included in the study. All the strains were preserved in sterile tubes with Sabouraud dextrose broth at -80˚C until use.
All the isolates were identified both by API ID32C (bioMerieux, France) and MALDI TOF (Vitek MS, bioMérieux, France) in addition to their morphological characteristics (9). Molecular analysis was also performed targeting IGS1 region. T. asahii CBS 2479, Candida krusei ATCC 6258 and C. parapsilosis ATCC 22019 standard strains were used as controls.
DNA isolation and sequencing
We extracted DNA from the isolates by using DiversiLab™ MoBio UltraClean™ Microbial DNA Isolation Kit (bioMѐrieux, France) and separated DNA fragments by using gel electrophoresis method. IGS1 gene product obtained using 26S forward ‘5′-ATCCTTTGCAGACGACTTGA-3’’ and 5S reverse ‘5′-AGCTTGACTTCGCAGATCGG-3’’ oligonucleotide primers were sequenced by Sanger sequencing method (10).
Genotyping and Phylogenetic Analysis
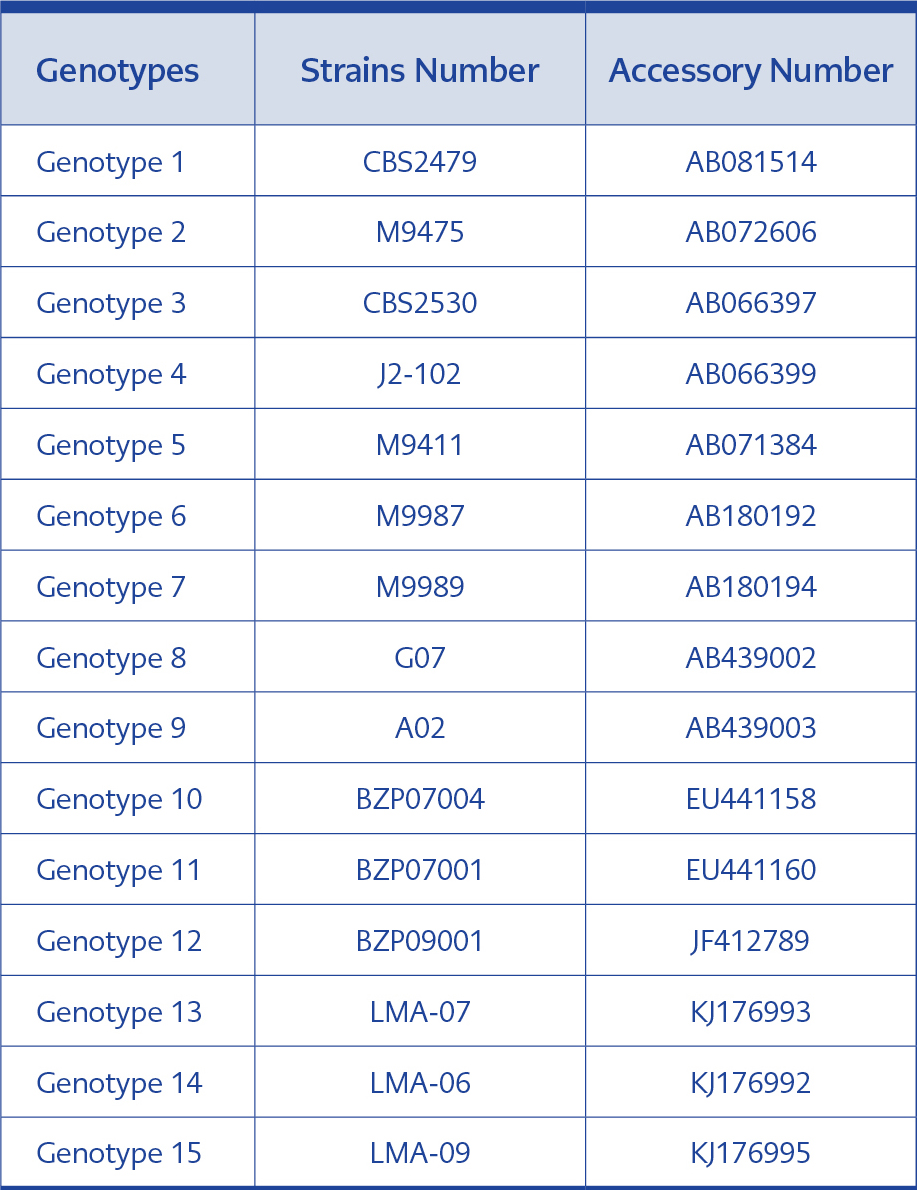
For genotyping, the IGS1 sequences of 70 clinical isolates and the sequences of T. asahii CBS 2479 and of 15 reference genotypes were aligned using BlastN programme, and the genotypes were detected by comparing the results (Table 1). The phylogenetic tree was generated using MEGA 7.01.21 programme. The statistical method UPGMA (Unweighted Pair Group Method with Arithmetic Mean) and bootstrap analysis with 1000 replications were used during phylogenetic tree construction. Kimura’s 2-parameter model was applied for calculating the genetic distances. Cryptococcus neoformans var. neoformans AJ300916 and C. neoformans var. gattii AJ300920 were used as the outgroups (16).
Antifungal susceptibility testing
Broth microdilution (BMD) method was performed to detect AMB (PAN Biotech, Germany), FCZ (Cayman, USA), VOR (Cayman, USA) and POS (Cayman, USA) susceptibility, and the results were evaluated according to CLSI M27-A3. The MICs for antifungals were read for AMB and azoles as the 100% and 50% inhibition of growth compared to controls, respectively.
In this study, all the practical experiments were performed in triplicate and repeated at least once if necessary.
Statistical analysis
Statistical analysis was performed by using the SPSS Statistic 23 software (IBM Corp, USA). As the number of strains was insufficient, genotype 3 and 5 were not analyzed.
The logarithmic values of all MICs (MIC GMs; MIC geometric means) were calculated and the significance of the difference between the GMs of genotype 1 and 4 was detected according to Mann- Whitney U test applied for the variables. The statistical significance was set as <0.05 (1).
Results
In total of 70 clinical specimens, urine samples showed the most significant growth [62] of T. asahii followed by blood [2], peritoneal fluid [2], bronchoalveolar lavage [2], sputum [1], and jejunum [1].
Genotyping and Phylogenetic Analysis of the Strains
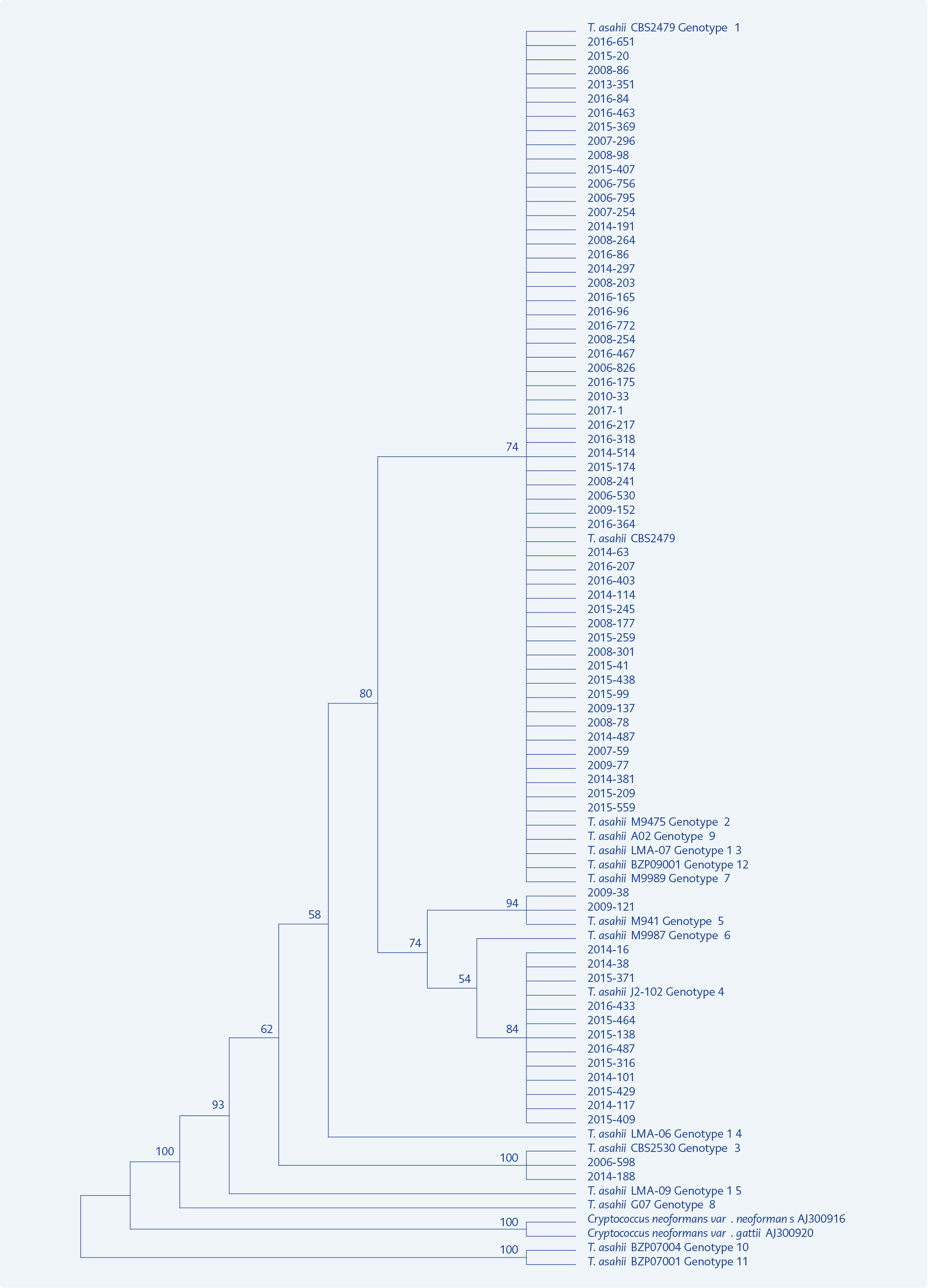
Figure 1. Phylogenetic tree constructed for distribution of 70 T. asahii clinical isolates and 15 reference genotypes.
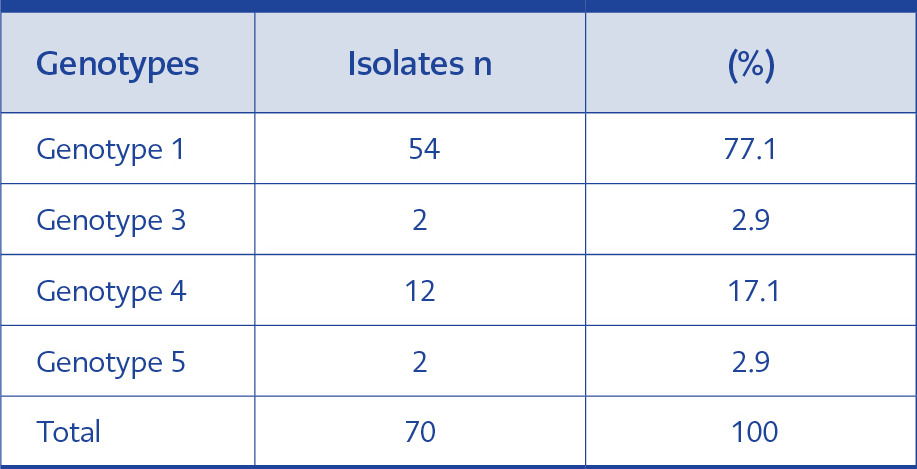
Four different genotypes (genotype 1, 3, 4 and 5) were detected in clinical isolates. We found that the majority of our isolates belonged to genotype 1 (77.1%). The genotype distribution of 70 isolates is shown in Table 2.
Distribution of genotypes varied according to years. Genotype 1 strains clustered between 2006- 2017 while genotype 4 was collected between 2014- 2016. Two strains of genotype 5 belonged to 2009, while genotype 3 was identified as one in 2006 and one in 2014.
In the phylogenetic tree constructed using MEGA 7.0.21 programme with 70 genotyped strains and reference genotypes, our 54 strains belonging to genotype 1 took place in the same branch, while 12 strains of genotype 4, two strains of genotype 3 and two strains of genotype 5 were clustered in separate branches. The reference genotypes 1, 2, 7, 9, 12 and 13 differed from each other only by one or two base pairs and were clustered in the same branch indicating close relationship between them (Figure 1).
Antifungal susceptibility tests
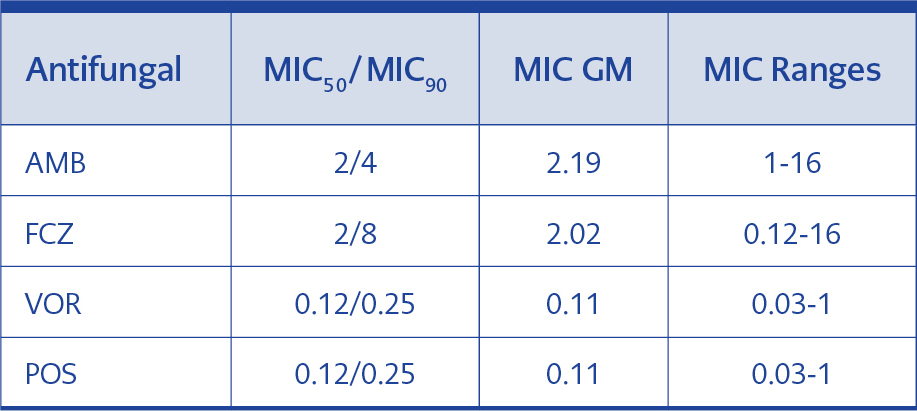
MIC50/MIC90, MIC GM and MIC range of AMB, FCZ, VOR and POS were determined to define their efficacy for 70 T. asahii isolates. MIC50/MIC90 and MIC GM values of AMB and FCZ for all strains were found to be higher than that of VOR and POS (Table 3).
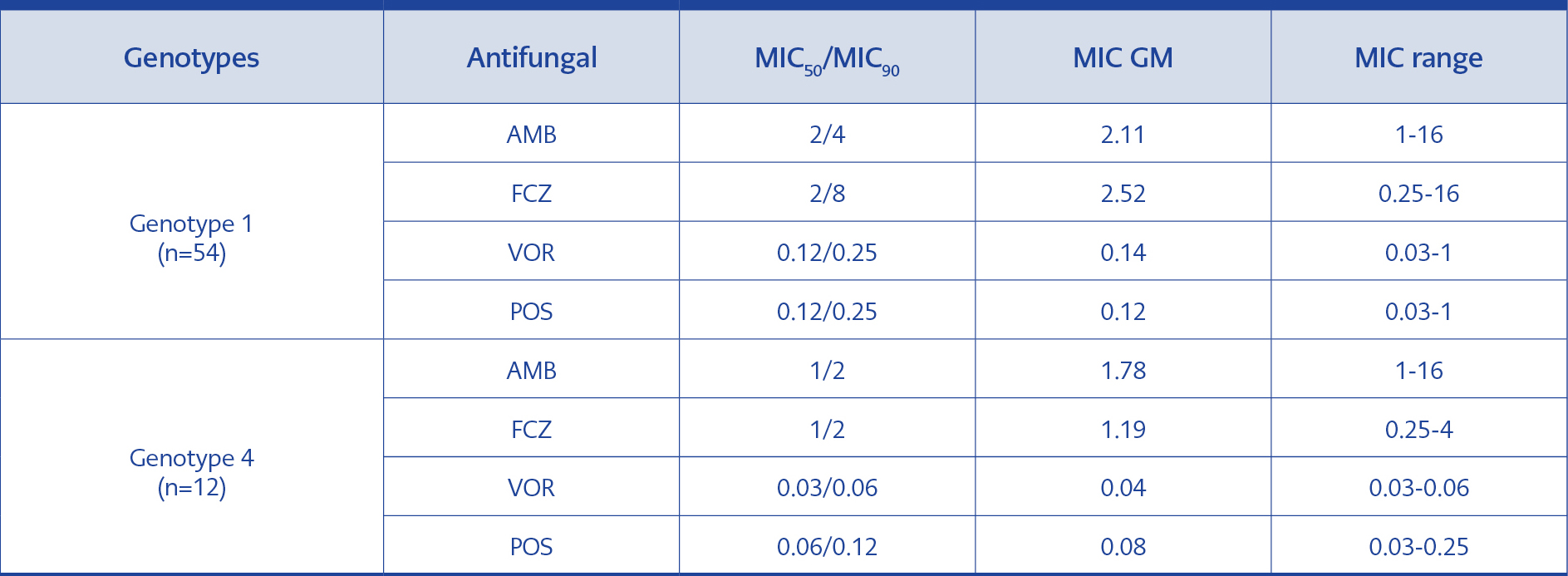
Table 4.MIC50/MIC90, MIC GM values and MIC ranges in the isolates of genotype 1 and genotype 4 (μg/ml).
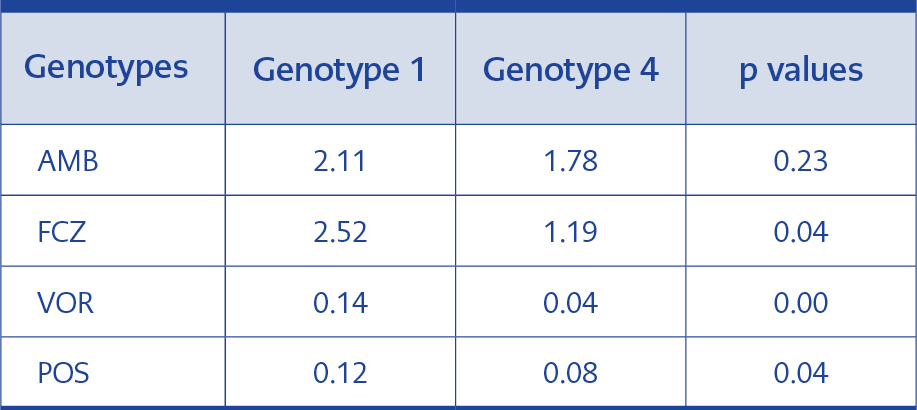
Table 5.Statistical analysis of MIC GMs of genotypes 1 and 4.
The analysis was performed on the basis of genotype MIC50/MIC90, MIC GM and MIC range values in genotype 1 were found to be higher than the 12 strains in genotype 4 with the exception of AMB MIC ranges that were equal between the two genotypes (Table 4).
Result of statistical analysis
The difference between the FCZ, VOR and POS MIC GMs for the isolates in genotype 1 and 4 was statistically significant, but no significant difference regarding AMB MIC GMs was detected (Table 5).
Discussion
Trichosporon asahii is an emerging fungus during 20 years (8). This fungus can lead to highly invasive infections with a high mortality rate, especially in immunocompromised patients. T. asahii exhibits primary resistance to echinocandin group of antifungals and flucytosine and AMB as monotherapy has no clinical efficacy (19).
Another important feature of T. asahii is its diverse genotypes according to different geographical regions. This diversity can lead differences in susceptibility profiles against antifungals and in virulence levels (16).
In this study, 70 T. asahii strains isolated from clinical samples of the patients admitted to Marmara University Hospital between 2006- 2017 were genotyped and their susceptibilities to various antifungals were investigated in order to yield epidemiological data.
Genotyping of T. asahii isolates has been carried out in order to determine the genotypic diversity in different geographical regions. So far, fifteen genotypes were detected in the world. In the study of Sellami et al., the authors detected 13 out of 28 (46.4%) T. asahii strains as genotype 1, and the following one was genotype 4 (35.7%) (16).
In the same study they also stated that genotype 1 with the proportion of 45% to 87% was the most common genotype within T. asahii isolates obtained from different countries including Japan (2002), Argentina (2007), Spain (2007), Brazil (2009), Turkey (2010) and Thailand (2010). They also analyzed the data from other countries and found that genotype 1 was never isolated in the United States in 2002, while the most frequently isolated one was genotype 3. According to the other studies from Argentina (2014) and India (2016), genotype 3 was the most common one. It was also reported that genotype 4 was the most widespread one in China (2011) and Greece (2014) (16).
Kalkancı et al. did the first research related to the diversity of genotypes of T. asahii in Turkey. The authors yielded mostly genotype 1 (79.3%) followed by genotype 5 (8%), genotype 3 (6.9%), genotype 4 (1.1%) and genotype 9 (1.1%). For the first time in the world, they obtained 87 isolates from 4 different geographical areas (10). In a recent study in Turkey by Hazırolan et al. (2019), almost all the findings were similar to that of Kalkancı et al.; the genotype distrubutions in 68 T. asahii strains have been reported as 79.3% genotype 1, 8% genotype 5, 6.9% genotype 3, 3.4% genotype 6, 1.1% genotype 4 and 1.1% genotype 9 (9).
Our findings were similar to that of Kalkancı et al., and Hazırolan et al., as genotype 1 (77.1%) was the most detected, and followed by genotypes 4 (17.1%), 3 and 5 (2.9% each) (Table 2).
Another goal of genotyping is to determine the source of outbreaks. In our study, it is seen that genotype 1 spreads in a wide range of years between 2006- 2017. Genotype 4 (n=12) clusters in the years 2016 and tended to increase between 2014- 2015 (Table 2). However, we cannot claim that this accumulation may show an outbreak since most of the strains were isolated from patients in different clinics.
We also examined the antifungal drug susceptibility of T. asahii isolates. Despite the increased frequency of the infections caused by this fungus, there are relatively fewer data on the susceptibility of T. asahii to antifungal agents when compared with the studies with Candida species in Turkey.
The MIC breakpoints are not defined for this species previously. However, in the study of Francisco et al. (2019), the authors proposed epidemiological cutoff values (ECV) for T. asahii for AMB (4 μg/ml), FCZ (8 μg/ml), VOR (0.25 μg/ml) and POS (0.5 μg/ml). Since these values have not been established yet, we did not use them for our isolates (6).
AMB has been used for many years in the treatment of systemic Trichosporon infections. However, in recent years it has been reported that even if the levels of AMB MICs are low, the drug cannot reach the serum level required for the fungicidal effect and the strains may develop tolerance against this agent (20, 21). In the study conducted in Turkey by Uzun et al., eight Trichosporon species exhibited sensitivity against AMB. In this study, the researchers determined MIC90 value as 1 with RPMI and as 0.125 μg/ml with AM3 medium (22). In 2002, Arikan et al. used the guidance of NCCLS M27-A2 and found that the MIC range of AMB was 1-8 μg/ml at 48th hour of incubation in 43 T. asahii strains (3). Metin et al. examined 27 strains of Trichosporon (13 were T. asahii) and found MIC90 for AMB as 1 μg/ml (13). Kalkancı et al. found that T. asahii isolates showed low sensitivity to AMB; as their 87 isolates exhibited MIC50/MIC90 of 1/2 and MIC GM of 1.138 µg/ml (10). However, in our study, 70 isolates were found to have MIC50/MIC90 of 2/4 and MIC GM of 2.186 µg/ml (Table 3).
In the studies of Chagas-Neto et al. (2009) in Brazil, Yang et al. (2013) in Taiwan and Montoya et al. (2015) in Mexico, the authors found that 47%, 33.3% and 53% of T. asahii isolates yielded high AMB MIC values (≥2 μg/ml), respectively, according to CLSI BMD method (4, 14, 23). In the recent study by Sellami et al., 82.1% of the 28 T. asahii strains had MIC values of ≥2 μg/ml (16). In our study, we found ≥2 µg/ml MIC values for AMB in 70% of the isolates, as well. Our results for AMB are similar to the findings of the studies above.
The in vivo and in vitro activities of triazoles against Trichosporon species are higher than that of AMB (5, 14). Trevino-Rangel et al. showed that new azoles such as POS and VOR for T. asahii are more effective than FCZ and AMB (21). Uzun et al. found that VOR was the most active drug against T. asahii (22). Metin et al. reported that MIC90 was 4μg/ml for FCZ while 0.125 μg/mL for VOR (13). In another study by Kalkanci et al., it has been emphasized that the efficacy of FCZ with MIC GM 13.66 was very low and the most active drug was VOR with 0.154 MIC GM (10). In the study of Hazirolan et al., the lowest MIC GM was obtained for VOR when compared with FCZ, POS, ITZ and isavuconazole (8).
In the studies in Brasil (Chagas-Neto et al. 2009) and USA (Pfaller et al. 2013), VOR was found to yield the lowest MIC value when compared with other azoles (4, 15). Similarly, in another study by Trevino et al. using the EUCAST guideline, MIC GMs of AMB, FCZ and VOR were 1.25; 5.30 and 0.31 respectively, and the most active drug was found to be VOR (20). In the study conducted in Tunisia, the MIC ranges for FCZ was 0.5-32 and was 0.016-0.38 μg/ml for VOR, indicating that the latter is more active than the first one (16).
When we evaluate our findings concerning antifungal susceptibility results of triazole group, the results for FCZ were relatively high with the MIC range of 0.12-16; MIC50/MIC90, of 2/8, and MIC GM of 2.019 while, the most active drugs were VOR and POS both of which exhibited MIC GM of 0.107 μg/ml (Table 3).
The similar results in Turkey and abroad may reflect that the FCZ may not be used in the treatment of T. asahii infections soon. Although some reports concern the use of fluconazole, the fact that in vitro MIC values for this antifungal reveals heterogeneous results, VOR is suggested as the first choice of therapy according to the clinical outcome of the patients (1,2).
There are several studies investigating the relationship between genotype and antifungal susceptibility profiles of T. asahii. In a study conducted in Thailand, no significant difference was found between antifungal MIC values of genotypes 1 and 3, which were the most frequent genotypes in 101 T. asahii strains (12). Another study, in Taiwan in 2013, reported that genotype 4 was more resistant to AMB, FCZ and VOR than the genotypes 1, 3, 5 and 7. However, the difference was not statiscally significant and it was suggested that more strains should have been investigated to detect whether there was a relationship between antifungal susceptibility profiles and genotypes (23).
Sensitivities for AMB, FCZ, VOR and caspofungin of 28 T. asahii strains was investigated in Tunisia, but no meaningful difference was found between genotypes 1, 3, 4 and 7 (p> 0.05) (16). In the review of Almeida Junior and Hennequin, they stated that there was no correlation between individual genotypes and sensitivities, but when combined the results of four different studies, they emphasized that VOR MIC values for genotype 3 strains were higher than genotype 1 strains, and this difference was statistically significant (1). Similarly, in the study of Guo et al. (2019), the authors found that genotype 3 strains exhibited higher AMB MICs than did genotype 1 and 4 strains (7).
In our study, genotype 1 and 4 were compared for antifungal MIC values (Table 4). Genotype 1 had higher MIC GMs for FCZ, VOR and POS than genotype 4, and these differences were statistically significant (p≤0.05). Nevertheless, our result reflects the fact that in terms of FCZ, genotype 4 may be more sensitive than genotype 1, albeit at a level not affecting the therapeutic efficacy of VOR and POS. No statistically significant difference was found between genotype 1 and 4 for AMB (p>0.05) (Table 5).
Conclusions
As a result, T. asahii isolates used in our study clustered in genotype 1 mostly, which had MIC values high for AMB and FCZ and very low for VOR and POS. Although we did not investigate the clinical situation of the patients, half of them stayed in intensive care, bone marrow transplantation and cardiovascular surgery units at which the patients took prophylactic antifungal therapy that might induce high MIC values in our isolates against AMB and FCZ. Besides, it is an outstanding finding that genotype 4, which has higher virulence characteristics than other genotypes, was isolated in an increasing frequency in our hospital in recent years. We need more observational studies about genotyping and antifungal susceptibility testing, searching their relationship to define appropriate treatment approaches for often fatal T. asahii infections and to emphasize central and widespread epidemiological data, particularly in light of the ongoing migrations from different geographic regions in our country and throughout the world.